Lecture 5: Evolutionary Algorithms
Evolution in Nature
Evolution is the change in inherited characteristics of biological populations over successive generations
- Heritable characteristics/ traits
- such as the colour of ones eyes
- passed from one generation to the next in DNA
- Change or genetic variation comes from:
- mutations: random changes in the DNA sequence
- Crossover: re-shuffling of genes through sexual reproduction and migration between populations.
The driving force of evolution is natural selection - the survival of the fittest
- Genetic variation that enhance the survival and reproduction become and remain more common in successive generations of a population.
How do we map the relationship between evolution and optimisation?
What are Evolutionary Algorithms?
Evolutionary Algorithms (EAs) are a subset of meta-heuristic algorithms inspired by biological evolution, which include: Genetic Algorithms, Evolutionary Programming, Evolution Strategies and Differential Evolution
They are essentially a kind of stochastic local search optimisation algorithm.
A characteristic of EAs is that they are population based. i.e. they generate, maintain and optimise a population of candidate solutions
X_0 := generate initial population of solutions terminationFlag := false t := 0 Evaluate the fitness of each individual in X_0 while (terminationFlag != true) { Selection: select parents from X_i based on their fitness Variation: Breed new individuals by applying variation operators to parents Fitness Calculation: Evaluate the fitness fo new individuals Reproduction: Generate population X_t++ by replacing least-fit individuals t ++ If a termination criterion is met: terminationFlag := true }
Building Blocks of Evolutionary Algorithms
An Evolutionary Algorithms is made up of:
- A representation each solution is called an individual
- A fitness (objective) function: to evaluate solutions
- Variation operators: mutation and crossover
- Selection and reproduction: survival of the fittest
Optimisation is about finding a global optimum. To do this we must balance exploration against exploitation
In order to use an EA to solve a problem you need each of the building blocks listed above
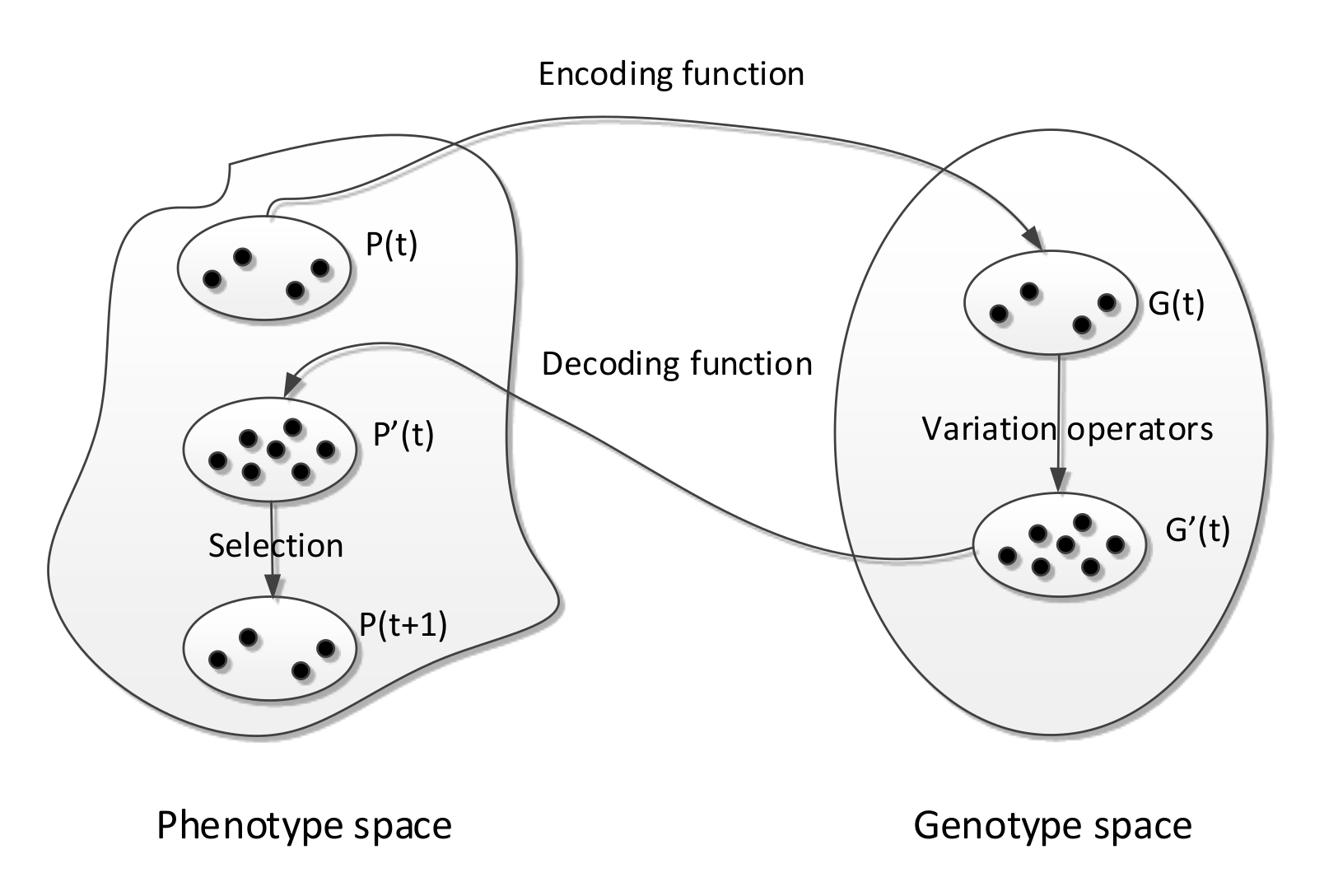
Representation
This is a way to represent or encode solutions
If we had a optimisation problem where the fitness of a solution is given by , where is the candidate solution.
- Solution is called a phenotype
- We encode the solutions using some form of representation
- The representation of a solution is called a genotype
- Variation operators act on genotypes
- Genotypes are decoded into phenotypes
- Phenotypes are evaluated using the fitness function
- Decoding and encoding functions map phenotypes and genotypes
- The search space of solutions is the set of genotypes and phenotypes
The selection of our representation depends on the problem.
Some possible representations include but are not limited to:
- Binary representation
- Real number representation
- Random key representation
- Permutation representation
Binary Representation
A traditionally very popular representation for use in genetic algorithms
Represent an individual as a bit string of length : - Genotypes
Map phenotypes to bit string genotypes via an encoding function
Map genotypes to phenotypes by a decoding function
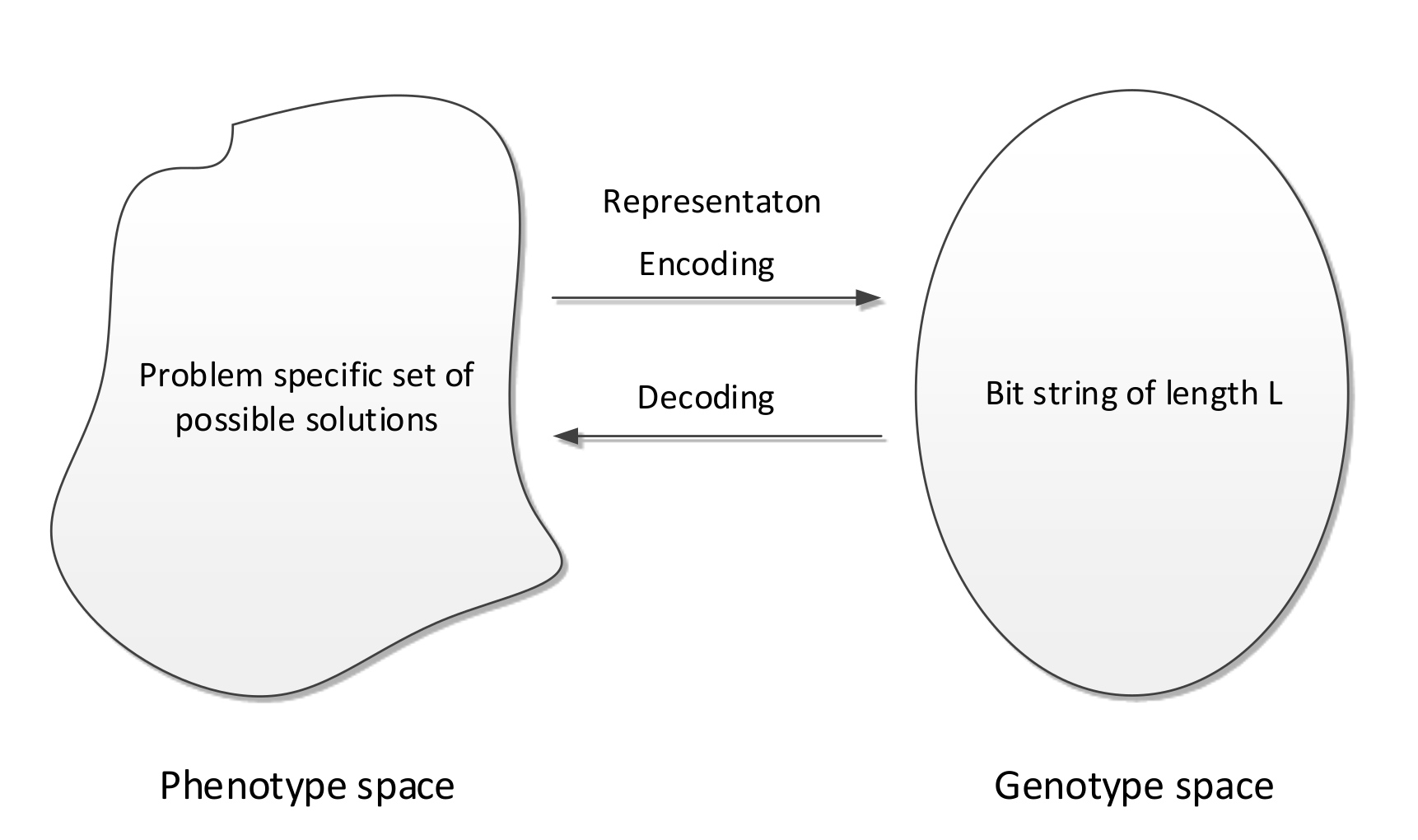
Decoding Function
Using a bit string to represent binary or integer solutions is trivial however, given an optimisation probelm with continuous variables, how do we represent them using a bit string of length
- Note: Usually each continuous variable will have an interval bound such as :
To solve this:
- Divide into segments of equal length
- Decode each segment into an integer and,
- Apply decoding function i.e. map the integer linearly into the interval bound :
Mutation
To mutate our candidate solutions we flip each bit with a probability of , called the mutation rate
A standard mutation rate is but can be in the range
If we use a low mutation rate, we can see this as creating a small a small random perturbation on the parent genotype. The mutated offspring are largely the same as their parents so will be close together in a search space relative to their Hamming distance.
Together with selection mutation performs a stochastic local search: it exploits the current best solutions by randomly exploring the search space around them.
Crossover
In the crossover process, two parents are selected randomly with a probability
In 1 point crossover: select a single crossover point on two strings, swap the data beyond that point in both strings
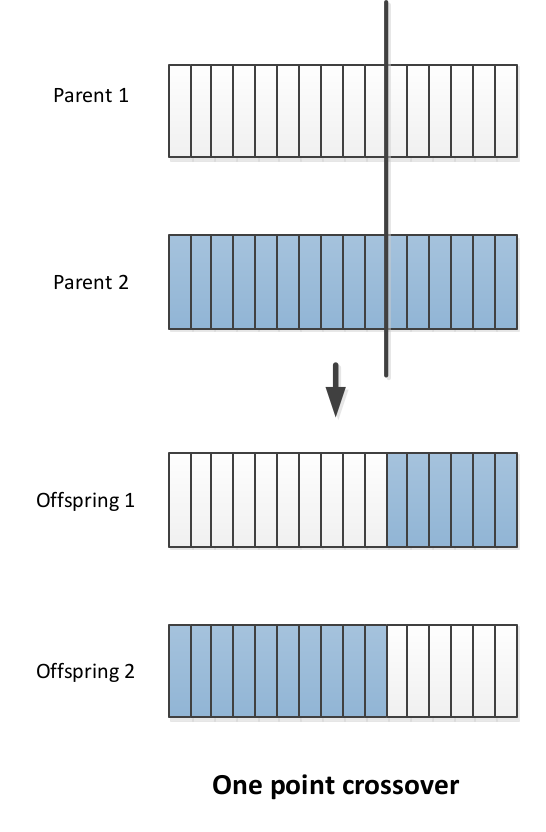
In point crossover:
- Select multiple crossover points on two strings,
- Split strings into parts using those points
- Alternating between the two parts and concatenate
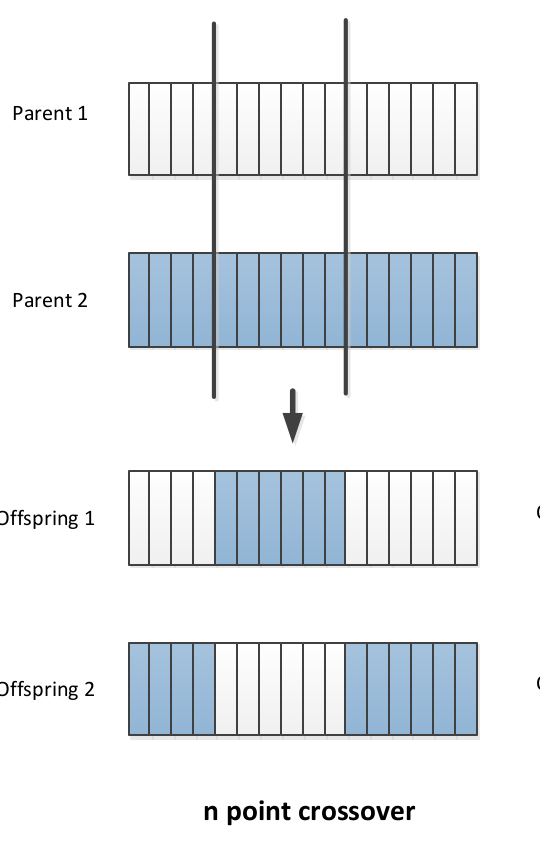
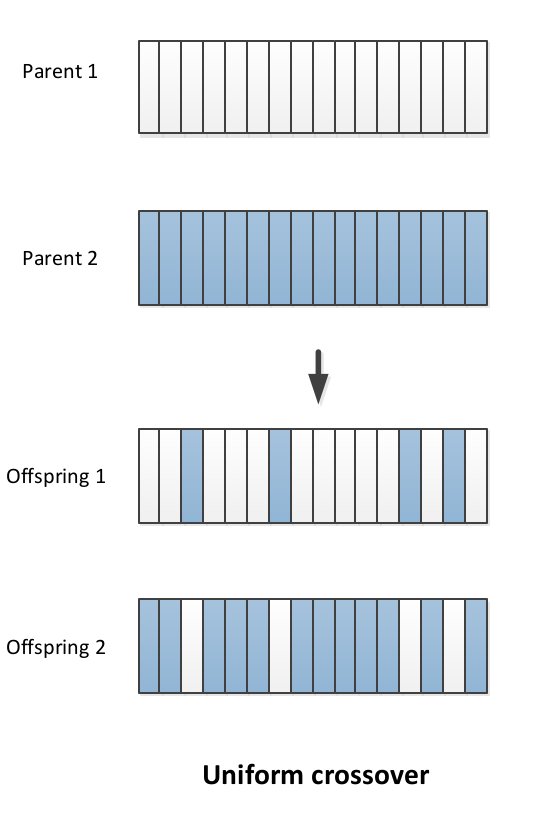
In Uniform crossover:
- , toss a coin
- If head: copy bit from parent 1 to offspring 1, parent 2 to offspring 2
- If tail: copy bit from parent 1 to offspring 2 and parent 2 to offspring 1.